Results
To investigate which transcription factors are involved in regulation of the gut cell lineage in sea urchin development, we used a novel approach to physically isolate the archenteron cells of Strongylocentrotus purpuratus embryos at 30 hours post-fertilization without perturbation of transcript representation and compared the difference in gene expression to distinguish archenteron-specific transcription factors. Figures and findings obtained using computer analysis software R. Data was normalized to gene length and the sum of all sequencing reads. The Cox-Reid profile-adjusted likelihood (CR) method was used to estimate dispersion. A likelihood ratio test was performed to test for differential expression using the edgeR package. This generated p-values for each gene, which were used to determine if difference in expression between the gut (archenteron) and all other cell populations was statistically significant.
Differential gene expression analysis in S. purpuratus embryos at 30 hours post-fertilization (hpf) revealed that, amongst 20,973 genes, 104 were enriched and 306 were depleted in the archenteron. Among these, a single transcription factor was identified amongst the enriched genes while all depleted transcription factors have unrelated function and were not included in subsequent analyses. Eight transcription factors from the list of non-differentially expressed genes were singled out as having potential function in archenteron regulation but their gene expression profiles revealed that their expression did not peak at 30 hpf. Results strongly suggest that FoxA alone is a pivotal transcription factor in the formation of the archenteron.

Figure 1.
Figure 1. Comparative transcriptome analysis in the S. purpuratus archenteron at 30 hours post-fertilization. (A) Volcano plot illustrating p-values and fold change obtained from a GLM likelihood ratio test used to test for differential gene expression. Thresholds for differentially expressed genes are depicted by the dotted lines (p<0.05; fold change>1). Archenteron cells are the cell type of interest while all other cells represent the control cell population. Data reflects consensus among two replicates. (B) Scatterplot illustrating the abundance of every mRNA species in archenteron cells relative to control, with data from replicate 1 (pink points) and 2 (light blue points) superimposed, demonstrating that replicates are concordant. FoxA (whose cis-regulatory apparatus was used to create the BAC to confer specific expression of GFP to archenteron cells) is labeled for both replicates. (C) Scatterplot illustrating the abundance of every mRNA species enriched to the archenteron cells (red points) relative to control. Blue points illustrate depleted transcripts. Note that colour codes reflect consensus among two replicates, whereas the individual data points shown represent the mean transcript abundance as observed for replicates 1 and 2. For all analyses, the number of sequencing reads per gene has been normalized according to gene length and the total count observed for all reads.

DE transcription factors of interest
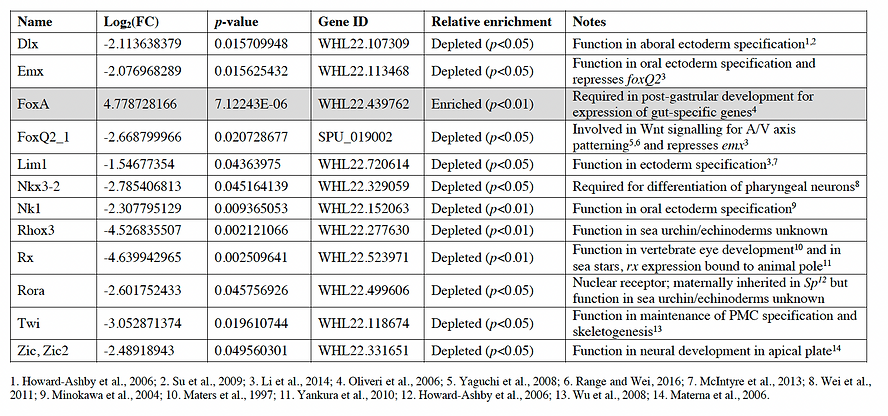
Table 1.
Table 1. Differentially expressed transcription factors in the archenteron of S. purpuratus embryos at 30 hours post-fertilization. Fold change (log2 transformed), p-values, gene ID, and relative enrichment of transcription factors that were differentially expressed in the archenteron of S. purpuratus embryos based on analysis from Fig. 2. FC=fold change.
FoxA | Upregulated in the gut | Class: TF
-
Oliveri et al., (2006): FoxA is expressed throughout the gut following gastrulation and is required in post-gastrular development for the expression of gut-specific genes (left).
-
Tu et al., (2006): “Genetic and functional studies have shown Fox TFs to be involved in the regulation of many biological processes. They play important roles in embryonic development, cell fate specification and cell differentiation, and morphogenesis, as well as in regulation of cell cycle and metabolism, and as effectors of signal transduction and chromatin structure (Carlsson and Mahlapuu, 2002, Pohl and Knochel, 2005).”
-
de-Leon & Davidson (2010): “…major function of the foxa gene during embryonic development is maintenance of the endoderm–mesoderm boundary, by repression of mesoderm fate in the endoderm. Interference with translation of foxa mRNA in sea urchin causes loss of gut formation and specification of excess mesodermal derivatives (Oliveri et al., 2006).”
-
Right: Four separate cis-regulatorymodules (CRMs) cooperate to control foxa expression in different spatial domains of the endomesoderm, and at different times… The complex and dynamic expression of foxa is regulated by a combination of repressors, a permissive switch, and multiple activators.
-
References
Rx (retinal homeobox) | Downregulated in the gut | Class: Nervous, TF
-
The transcription factor Rx plays an important role in vertebrate eye development (Mathers et al, 1997). Rx regulates expression of pax6 and six3 during early eye development, and the expression of opsin and other effector genes in differentiated ciliary photoreceptors in vertebrates (Nelson et al., 2009, Zhang et al., 2000, Zuber et al., 2003, Pan et al., 2010).
-
Yankura et al. (2010): In the sea star (P. miniata), the animal pole has a distinct regulatory state defined by foxq2, pax6, pea3, zic, rx and six3 expression. Transcripts of rx are only found in the animal-most ectoderm in blastulae and expression is maintained in the animal pole of gastrulae (top left).
Yankura et al. (2010)
Rhox3 (retinal homeobox Rx1) | Downregulated in the gut | Class: TF
-
96 homeobox genes in S. purpuratus (Howard-Ashby et al., 2006).
-
Similar to Rx (above)
Twi (twist) | Downregulated in the gut | Class: TF
-
Wu et al., (2008): Twist expression is required for the maintenance of the PMC specification state, and a reciprocal regulation between Alx1 and Twist offers stability for the subsequent processes, such as PMC differentiation and skeletogenesis.
-
Top: In L. variegatus (B–G) WMISH shows the expression domains of Lvtwist mRNA. Lvtwist mRNA is detected in the vegetal plate (C), the early ingressing PMCs (arrow in panel D) and ingressed PMCs (E). (F) Early gastrula stage (EG): Lvtwist mRNA starts to be expressed in SMCs (arrow) and also persists in PMCs. (G) Late gastrula stage (LG): Lvtwist mRNA continues to be expressed in PMCs (arrowhead), the SMC territory (arrow) and the archenteron.
-
Bottom: Fig. 8. The pre-EMT and post-EMT regulatory states in the PMC GRN. (A) The PMC GRN before ingression. Alx1, Ets1, Tbr and Delta are all regulated by de-repression of Pmar1, which is activated by the nuclear β-catenin. Pmar1 represses HesC allowing activation of the early micromere GRN (shown faded-out as events at earlier stages). Alx1 positively regulates Twi, Dri, and Sna. Ets1 also has a positive input on Dri. Both Twi and Dri activate downstream differentiation batteries (Msp130, Sm50). Snail controls PMC ingression through regulating Cadherin expression. Delta signals to adjacent veg2 cells through Notch to specify SMCs (not shown). (B) After PMC ingression is completed, the expression of Sna and Dri is downregulated from PMCs (by an unknown repression mechanism), while Alx1, Twi and other genes (not shown) continue to maintain the expression of PMC differentiation genes for the skeletogenesis at later stages. Twi feeds back to Alx1 to maintain its expression, and Ets1 also regulates Alx1 at this time. However, it is still uncertain if Alx1 regulates Twi at this stage. The Alx1–Twi reciprocal positive regulation may provide stabilities for the PMC GRN during PMC ingression.
-
Alx1 is a positive regulator upstream of twist, and Twist is involved in the maintenance of alx1 action = positive feedback loop
-
It has been shown that Drosophila WntD is feedback inhibitor of the Dorsal/Twist/Snail network in the gastrulating embryo (Ganguly et al., 2005, Gordon et al., 2005). Thus in the sea urchin embryo, it is possible that a similar inhibiting signal or a transcriptional repressor is activated immediately after PMCs ingress and represses genes, including snail, which are not required in the post-ingression PMC GRN.
-
Nkx3-2 (NK3 homeobox 2) | Downregulated in the gut | Class: TF
-
Nkx homeodomain factors are a family of homeodomain TFs that are critical regulators of organ development. Nkx3-2 plays a role in axial and limb skeleton development in mammals (Lettice et al., 2001)
-
Wei et al. (2011): Six3 (a TF) supports expression of Nkx3-2, which is required for differentiation of pharyngeal neurons in the oral animal pole ectoderm and the foregut.
-
Six3, which is required for these foregut neurons and all embryonic neurons, is transiently expressed in the foregut lineage. The Six3-dependent gene, Nkx3-2, which is expressed only in the oral anterior ectoderm and the foregut, is also essential for the development of pharyngeal neurons.
-
Top: Nkx3-2 is expressed in oral animal pole ectoderm and foregut endoderm during gastrulation (C) but not in Six3 mutants.
-
-
Wei et al. (2016): Specification of pharyngeal neurons occurs through a neurogenic pathway that includes Six3 and Nkx3-2 (Wei et al., 2011), which also includes SoxC and Brn1/2/4.
-
Bottom: Model for the neurogenic pathways. SoxC was previously identified as being dependent on Six3. SoxC expression is activated in isolated neural precursors by unidentified factors and cell signaling mechanisms (dotted arrow). Cells expressing SoxC are dividing (circular arrow) and at least some express Delta and are subject to lateral inhibition. SoxC is required for the development of neurons expressing serotonin and Tph (upper pathway) and also for all other neurons expressing SynB (lower pathway). SoxC is required for expression of Brn1/2/4 in both pathways; SoxC is transiently expressed and some cells are observed to contain both mRNAs. Brn1/2/4 also is transiently expressed, and in later embryos some Brn1/2/4-expressing cells also express the differentiated marker SynB. Z167 is expressed only in developing serotonergic neurons (upper pathway) and is essential for their development. Its position in the hierarchy relative to Brn1/2/4 remains to be defined, but Z167 is likely to function in parallel or downstream.
-
The requirement for several genes has been demonstrated by functional assays, including Zfhx for serotonergic neurons (Yaguchi et al., 2012) and Nkx3.2 for pharyngeal neurons (Wei et al., 2009)
-
References
Wei et al. (2011)
Wei et al. (2016)
FoxQ2_1 | Downregulated in the gut | Class: Nervous, TF
-
Yaguchi et al. (2008): Restriction of FoxQ2 to the animal plate is a crucial element of canonical Wnt signaling that coordinates patterning along the AV axis with the initiation of oral-aboral (OA) specification.
-
Top: "...foxq2 transcripts accumulate throughout the animal hemisphere of a 5th cleavage-stage embryo and then rapidly disappear from all the ectoderm except the animal plate during the next several cell divisions, in a process requiring nuclear β-catenin function. If this restriction is prevented, then FoxQ2 persists throughout the animal hemisphere and suppresses nodal transcription in the presumptive oral ectoderm. Importantly, clearance of FoxQ2 is the only nuclear β-catenin-dependent event that is required for maintaining nodal expression in oral ectoderm. Thus, instead of providing a positive input to nodal transcription, canonical Wnt signaling regulates nodal expression and establishment of OA polarity through a double repression mechanism—the elimination of a nodal repressor."
-
-
Range and Wei (2016): During early development, the sea urchin anterior neuroectoderm (ANE) territory separates into inner and outer regulatory domains that express the cardinal ANE transcriptional regulators FoxQ2 and Six3, respectively. FoxQ2 drives this patterning process, which is required to eliminate six3 expression from the inner domain and activate the expression of Dkk3 and sFRP1/5, two secreted Wnt modulators.
-
Bottom: extended model for sea urchin ANE restriction that integrates the FoxQ2, sFRP1/5 and Dkk3 signaling center (see main text for details). nβ-catenin, nuclear β-catenin.
-
References
Range and Wei (2016)
Yaguchi et al. (2008)
Rora (nuclear receptor ROR alpha) | Downregulated in the gut | Class: TF
-
The protein encoded by this gene is a member of the NR1 subfamily of nuclear hormone receptors. It can bind as a monomer or as a homodimer to hormone response elements upstream of several genes to enhance the expression of those genes. The encoded protein has been shown to interact with NM23-2, a nucleoside diphosphate kinase involved in organogenesis and differentiation, as well as with NM23-1, the product of a tumor metastasis suppressor candidate gene. Also, it has been shown to aid in the transcriptional regulation of some genes involved in circadian rhythm. Four transcript variants encoding different isoforms have been described for this gene (NCBI).
-
Specific association with ROR elements is required for ROR-alpha to function as a transcriptional activator.
-
-
Howard-Ashby et al. (2006); Figure 9: Sp-ror is maternally inherited and expression rises to within the minimum range estimated to be significant (150–350 copies/embryo) by 6 hpf.
References
Zic/Zic2 | Downregulated in the gut | Class: TF
-
Aruga (2004): Zic proteins are important regulators of neural development and interact with these
-
Materna et al. (2006): The sea urchin zic orthologue begins to be expressed at around 18 h postfertilization, and its transcripts are localized in the neurogenic apical plate (right).
-
In humans, acts as a transcriptional activator. Involved in neurogenesis. Important roles in early stage organogenesis of the CNS, during dorsal spinal cord development and maturation of the cerebellum (UniProt).
Materna et al. (2006)
Nk1 | Downregulated in the gut | Class: TF
-
Minokawa et al. (2004); Left (Fig. 6): No obvious expression of SpNk1 is observed in the embryos before 18 h (data not shown for WMISH, also see Fig. 6E). SpNk1 transcripts start to accumulate at around 24 h in mesenchyme blastulae (Fig. 6E). A subequatorial expression pattern is obvious by 24 h, confined to one side of the embryo (data not shown). This expression becomes stronger during gastrulation (Fig. 6A, and B), peaking at 36 h (Fig. 6E). The expression is restricted to the subequatorial oral ectoderm, including the lip of the blastopore on the oral side (Fig. 6A). This same pattern of expression is preserved in late gastrula (Fig. 6C) and pluteus stage larvae (Fig. 6D).
-
McIntyre et al. (2013); Right (Fig. 7): Short-range Wnt5 signal from the endoderm actively patterns the adjacent boundary ectoderm. This signal activates a unique subcircuit of the ectoderm gene regulatory network, including the transcription factors IrxA, Nk1, Pax2/5/8 and Lim1, which are ultimately restricted to subregions of the border ectoderm (BE).
References
Minokawa et al. (2004)

McIntyre et al. (2013)
Dlx (distal-less homeobox) | Downregulated in the gut | Class: TF
-
Howard-Ashby et al. (2006); Top (Fig. 6): Dlx expression is localized apically, aboral ectoderm. Important activator of the aboral ectoderm GRN.
-
Su et al., (2009); Bottom (Fig. 4): Dlx is expressed specifically in the aboral ectoderm and are linked into the GRN that specifies it there.
-
Four homeodomain genes, irxA, lhx2.9, dlx, and hox7, are all predicted to be locked together in feedback relations and irxA feeds back on tbx2/3, the first to be expressed, as well.
-
References
Howard-Ashby et al. (2006)
Su et al. (2009)
Emx | Downregulated in the gut | Class: Nervous, TF
-
EMX-1 is a member of the EMX family of putative transcription factors (Simeone et al., 1992) and plays a role in the specification of positional identity and the proliferation of neural stem cells, although the precise role of EMX-1 in embryonic development is not fully understood.
-
Li et al. (2014): The emx gene repressively controls the lower boundary of foxq2 expression after the midblastula stage. foxq2 expression is required to prevent expression within the apical domain of emx (and also of univin and egr) (Fig. S3A). Thus, emx and foxq2 are linked in a mutual exclusion circuit, in which, within the normal domain of expression of each, expression of the other is prohibited. Note that, later in blastulation, expression of emx, univin, and egr is cleared from the oral and/or aboral ectoderm (Fig. S1A) by repressors active in that domain, whereas their boundaries with foxq2 persist on the oral side of the embryo.
-
The apical neurogenic/ oral ectoderm boundary is formed with the aid of a second widely expressed soxB1 target gene, the repressor emx, required in the near-apical oral ectoderm where nodal and not expression are weaker. These repressions confine foxq2 expression to the apical domain. Then the exclusion functions kick in: foxq2 represses emx, and foxq2 plus six3 repress nodal and consequently the whole oral ectoderm GRN within the apical domain. The neurogenic foxq2 region is thus permanently segregated.
-
Li et al. (2014)
Lim1 | Downregulated in the gut | Class: Nervous, TF
-
Kawasaki et al. (1999): HpLim1 mRNA was localized in the vegetal plates of hatched blastulae, but it was not detectable in primary mesenchymecells (PMC) ingressed into the blastocele. While short-term expression of HpLim1 in the vegetalplate is needed for differentiation of aboral ectoderm, endoderm and PMC, ectopic expression of HpLim1 suppresses normal differentiation directing all embryonic cells to differentiate into oral ectoderm.
-
Li et al. (2014); see above right image: Within the oral ectoderm, another boundary forms—that separating the near-apical from the central oral ectoderm. Here another early regulatory player is six3, activated almost as early as is foxq2 in the animal hemisphere, and by 18 h is expressed in the upper half thereof. The repressive target ofsix3 plusfoxq2 islim1, which because it also is driven by SoxB1 could express throughout the ectoderm, but because six3 and foxq2 are expressed first, lim1 can be transcribed only up to the lower boundary of the six3 expression domain, defining a central oral ectoderm region, whereas the six3-expressing region between the foxq2 and lim1 boundaries becomes the near-apical ectoderm. Thereafter, a further exclusion is instituted: foxq2 excludes six3 expression from the central apical plate and confines its expression domain to the surrounding near-apical ectoderm.
-
McIntyre et al. (2013); Right (Fig. 7): Short-range Wnt5 signal from the endoderm actively patterns the adjacent boundary ectoderm. This signal activates a unique subcircuit of the ectoderm gene regulatory network, including the transcription factors IrxA, Nk1, Pax2/5/8 and Lim1, which are ultimately restricted to subregions of the border ectoderm (BE).
References
Other transcription factors
Looking at the data from replicates separately, I identified 19 TFs that, although not statistically significant given my analysis, could still plausibly be of interest. 16 of them appeared to be enriched in the archenteron at first glance, but the p-values fell above 0.05 and were therefore not recognized in our statistical analysis. The other three appear to be enriched in replicate 1 but not in replicate 2.
Table 2.
Table 2. Additional transcription factors of interest. Fold change (log2 transformed), p-values, gene ID, and mRNA abundance in GFP+ and GFP- populations in both replicates (measured in number of sequencing reads per gene) of transcription factors whose expression was statistically ambiguous but could plausibly be of interest. Highlighted rows indicate the eight transcription factors that may have a potential function in the archenteron of S. purpuratus embryos based on literature search. FC=fold change.


Alx4 | Class: TF | May be involved in coelom development
-
The echinoderm alx1 and alx4 genes, which are physically adjacent to one another, originated from a gene duplication event (Koga et al., 2016).
-
The function of Alx4 in early sea urchin embryonic development has not been tested experimentally.
-
Alx1 and Alx4 have similarities in amino acid sequences, have overlapping expression in PMCs, and alx4 is positively regulated by alx1 (Rafiq et al., 2014). Alx1 and Alx4, however, are not functionally interchangeable (Khor and Ettensohn, 2017).
-
In euechinoids, alx4 is expressed by PMCs but also by non-skeletogenic mesoderm cells (probably presumptive coelomic pouch cells) at the tip of the archenteron (Koga et al., 2016; Rafiq et al., 2012). The function of Alx4 has not been explored experimentally; however, it has been proposed that because the alx4-like gene in hemichordates is expressed in the coelomic mesoderm, the primary function of the ancestral gene was to support coelom development, a function which may be retained by alx4 in echinoderms (Koga et al., 2016)
References
Koga et al. (2016); Right (Fig. 2): The expression pattern of sea urchin Alx4. Red fluorescent signals indicate HpCalx (Alx4) transcripts and blue signals denote nuclei stained by DAPI. (O) Expression was evident in ingressed primary mesenchyme cells (PMCs) of the blastula mesenchyme. (P) In late-stage gastrulae, signals disappeared from PMCs but were observed in secondary mesenchyme cells (SMCs) located around the tip of the archenteron. (Q) Expression was restricted to the coelomic pouches of prism larvae.
Ash2 | set1/Ash2 histone methyltransferase complex subunit ASH2 | Class: TF
-
Transcriptional regulator (PubMed:12670868).
-
Component or associated component of some histone methyltransferase complexes which regulates transcription through recruitment of those complexes to gene promoters (PubMed:19131338).
-
Component of the Set1/Ash2 histone methyltransferase (HMT) complex, a complex that specifically methylates 'Lys-4' of histone H3, but not if the neighboring 'Lys-9' residue is already methylated (PubMed:19556245).
-
Little to no information about role and function in sea urchins/echinoderms available.
References
Cic | Capicua S. purpuratus homolog | Class: TF | Transcriptional repressor
-
Member of the high mobility group (HMG)-box superfamily of transcriptional repressors.
-
Studies suggest that the N-terminal region of this protein interacts with Atxn1 (GeneID:6310), to form a transcription repressor complex, and in vitro studies suggest that polyglutamine-expansion of ATXN1 may alter the repressor activity of this complex.
-
Little to no information about role and function in sea urchins/echinoderms available.
References
Cutl | Cut like homeobox 1 S. purpuratus homolog | Class: TF | Transcriptional repressor
-
The protein encoded by this gene is a member of the homeodomain family of DNA binding proteins. It may regulate gene expression, morphogenesis, and differentiation and it may also play a role in the cell cycle progression.
-
CUX1 belongs to the homeodomain (HD) transcription factor family, which was first identified as a sea urchin transcription repressor of the sperm H2B gene by binding to promoter element–CCAAT and competing the binding of other transcriptional activators (Barberis et al., 1987).
-
Little to no information about role and function in sea urchins/echinoderms available.
Dri | Deadringer | Class: TF | Skeletogenic fate regulator
-
Deadringer (dri) and foxb genes, which together with ets1 and alx1, and the (at this stage) ubiquitously active regulatory gene hnf6 (Otim et al., 2004), in various combinations directly regulate the diverse skeletogenic differentiation genes.
-
Amore and Davidson (2006); Top: The ETS-DRI skeletogenic gene battery control expression of cyclophilin, one of the differentiation genes activated in the skeletogenic territory as a terminal function of the endomesodermal gene regulatory network.
-
Amore et al. (2003): Spdri is a key player in two separate developmental gene regulatory networks (GRNs). Spdri is expressed in a biphasic manner, first, after 12 h and until ingression in the skeletogenic descendants of the large micromeres; second, after about 20 h in the oral ectoderm, where its transcripts remain present at 30–50 mRNA molecules/cell far into development. In both territories, the periods of Spdri expression follow prior territorial specification events.
-
Bottom (Fig. 2): Spatial and temporal expression of Spdri gene during embryogenesis. (A) Accumulation of Spdri RNA in pmcs resident in the vegetal plate of a prehatching blastula; (B) Spdri transcripts in pmcs as they ingress at early mesenchyme blastula stage; (C) Spdri expression begins in oral ectoderm in late mesenchyme blastula; no further expression in completely ingressed pmcs; (D) Strong expression throughout oral ectoderm at prism stage; (E) Expression confined to oral ectoderm, vegetal view. (F) Strong expression seen throughout oral ectoderm and the ciliary bands at pluteus stage, at the borders between the oral and aboral ectoderm
-
References
Amore and Davidson (2006)
Amore et al. (2003)
Fog | aka ZFPM1 | Class: TF | GATA cofactor
-
Fog is a multiple zinc finger protein and Gata co-factor. FOG and GATA-1 synergistically activate transcription from a hematopoietic-specific regulatory region and cooperate during both erythroid and megakaryocytic cell differentiation (Tsang et al., 1997).
-
Kiyama and Klein (2007) identified an S. purpuratus fog ortholog, spfog1, and showed that SpGataE and SpFog1 physically interacted. Spfog1 transcripts were maximal by early blastula stage but continued thereafter to be expressed at low levels. Knockdown of spfog1 using antisense morpholino oligonucleotides did not produce notable effects on endomesoderm specification. They hypothesize that spfog1 expression is distributed evenly throughout the embryo, but could not confirm this with in situ hybridization.
-
Although they demonstrated that SpGataE and SpFog1 could physically interact, we were unable to reveal a role for SpFog1 by morpholino knockdown. Thus, the function of SpFog1 is uncertain. SpFog1 may modulate SpGataC or SpGataE in subtle ways that would be difficult to detect by our analysis of knockdown embryos. Our expression analysis indicated that spfog1 transcripts are quite rare during embryogenesis. It may be that SpFog1 transcripts are expressed at higher levels during oogenesis, larval development, or after metamorphosis, and that SpFog1 modulates SpGataC or SpGataE at these other stages in the sea urchin life cycle.
-
References
FoxN2/3 | aka ZFPM1 | Class: TF | Required for normal PMC ingression
-
Rho and McClay (2011):
-
Expression of foxN2/3 mRNA begins in micromeres at the hatched blastula stage and then is lost from micromeres at the mesenchyme blastula stage. foxN2/3 expression then shifts to the non-skeletogenic mesoderm and, later, to the endoderm.
-
Pmar1, Ets1 and Tbr are necessary for activation of foxN2/3 in micromeres.
-
FoxN2/3 is necessary for normal PMC ingression, for expression of several skeletal matrix genes, for preventing transfating and for fusion of the PMC syncytium.
-
Right (Fig. 1): In situ hybridization of foxN2/3 expression in L. variegatus embryos
-
Rho and McCLay (2011)
GataC and GataL/GataE | Class: TF
-
Materna et al. (2013): GataE is a direct Gcm target and part of a feedback loop locking down the aboral regulatory state.
-
The most upstream gene of the hierarchical aboral non-skeletogenic mesenchyme (NSM) GRN is gcm, which is activated as a direct target of Notch signaling (Ransick and Davidson, 2006), in response to presentation of the Delta ligand by the adjacent skeletogenic mesoderm (SM) (Materna and Davidson, 2012, Revilla-i-Domingo et al., 2007, Sweet et al., 2002)
-
The first transcription factor genes specific to the oral NSM, prox1 and gataC, are transcriptionally activated in this region between 17 and 18 hpf
-
Pertubation of gataC doesn't affect any genes pre-gastrulation
-
Top (Fig. 1): Summary of gene expression profiles in NSM between 10 hpf and 25 hpf. Differentiation genes in italics.
-
Middle (Fig. 7): GRN for establishment and maintenance of the aboral NSM regulatory state. (A) Prior to NSM subdivision Delta signaling emanating from the skeletogenic mesoderm (SM) activates gcm. (B) Following separation of NSM segments, the aboral NSM regulatory state is locked down through positive feedback between gataE, six1/2–eya and gcm. This subcircuit is thus independent of the initial Delta input. Repression of delta is relieved after clearance of HesC.
-
-
Kiyama and Klein (2007): The authors provided substantial supporting evidence that SpGataE represses spec2a in the endomesoderm, but, although Gata proteins often associated with Fog co-regulators, it does so independently of SpFog1 expression. The authors hypothesize that SpFog1 modulates SpGataE and SpGataC at other stages of the sea urchin life cycle.
-
Bottom (Fig. 1b): Spatiotemporal expression pattern of SpGataE during S. purpuratus embryogenesis. Green represents anti-SpGataE labeling, and red represents nuclei labeled with propidium iodide... supporting the notion that the SpGataEexpressing cells were Veg2 progenitors of secondary mesenchyme and foregut (Fig. 1b—B2). By the midgastrula stage and subsequent late gastrula and pluteus stages, expression of SpGataE had expanded to include the mid- and hindgut as well as the secondary mesenchyme
-
References
Materna et al. (2013)
Kiyama and Klein (2007)
Hhex | Hematopoietically expressed homeobox | Class: TF
-
Ettensohn (2009): Hhex is a regulatory component of the skeletogenic centers of the embryo and acts in a network to engage in mutual, positive interactions that probably stabilize the system, and eventually activate terminal differentiation genes (biomineralization genes), often via `feed-forward' interactions.
-
Left (Fig. 5): Selected regulatory interactions among early specification genes and late transcriptional regulators in the micromere-PMC GRN. Yellow boxes indicate early specification genes and red boxes indicate late transcriptional regulators. Two of the early specification genes shown (ets1 and tbr) are also expressed maternally (indicated by stippling)
-
-
Materna et al. (2013): Once skeletogenic mesenchyme (SM) ingression into the blastocoel is complete, three genes that are first expressed in the SM, erg, hex, and ets1/2.
-
Right (Fig. 1) Once expressed in the NSM, erg, hex, and ets1/2 expression is restricted to the oral side.
-
References
Ettensohn (2009)
Materna et al. (2013)
Krl | Class: TF, ZNF
-
Davidson et al. (2002);
-
Left (Fig. 3): Krl acts as a transcriptional repressor of the soxb1 gene in the endomesodermal domain in which the krl gene is active following beta-catenin nuclearization (Howard et al., 2001).
-
-
Howard et al. (2001); Fig. 3: SpKrl is activated upon nuclear entry of β catenin. Embryos in which SpKrl translation is inhibited with MASO lack endoderm. Conversely, SpKrl mRNA injection rescues some vegetal structures in β-catenin deficient embryos. SpKrl negatively regulates expression of the animalizing transcription factor, SpSoxB1. We propose that SpKrl functions in patterning the vegetal domain by suppressing animal regulatory activities.
-
Right (Fig. 3): SpKrl signal was first detectable at the 60-cell stage when it is concentrated on one side of the embryo. The SpKrl expression pattern is dynamic at later developmental stages. By ~180-cell stage, the region of highest SpKrl mRNA concentration includes presumptive endoderm (Fig. 3C). By the hatching blastula stage (18 hours), the SpKrl mRNA distribution has modulated to a torus about the vegetal pole (Fig. 3D).
-
References
Davidson et al. (2002)
Howard et al.
(2001)
Lbx | Class: TF
-
This gene and the orthologous mouse gene were found by their homology to the Drosophila lady bird early and late homeobox genes. In the mouse, this gene is a key regulator of muscle precursor cell migration and is required for the acquisition of dorsal identities of forelimb muscles (NCBI).
Little to no information about role and function in sea urchins/echinoderms available.
References
Nr1m2 | Nuclear receptor | Class: TF
-
Miglioli et al. (2021): Nuclear Receptors (NRs) are a superfamily of transcription factors specific to metazoans that have the unique ability to directly translate the message of a signaling molecule into a transcriptional response. In vertebrates, NRs are pivotal players in countless processes of both embryonic and adult physiology, with embryonic development being one of the most dynamic periods of NR activity. Accumulating evidence suggests that NR signaling is also a major regulator of development in marine invertebrates, although ligands and transactivation dynamics are not necessarily conserved with respect to vertebrates
-
Little to no information about role and function in sea urchins/echinoderms available.
Pax6_1 | Class: Nervous, TF
-
Lesser et al. (2011): In the green sea urchin, the most distal portion of these tube feet contain five ossicles arranged as a light collector with its concave surface facing towards the ambient light. These ossicles are perforated and lined with pigment cells that express a PAX6 protein that is universally involved in the development of eyes and sensory organs in other bilaterians.
-
Zhao et al. (2015); Top: The expression of SIPax6 generally increased from the blastula stage to the eight-arm larvae. Like the expression patterns of SIOpsin4 and SIOpsin5, SIPax6 significantly increased its expression from the eight-arm larvae stage to juveniles.
-
Martik and McClay (2015): In many embryonic systems, cells home, or migrate in a directed fashion, from their place of origin to a distant target site. The migration of the small micromeres to the coelomic pouches (or "homing") in the sea urchin embryo provides an exceptional model for understanding the genomic regulatory control of morphogenesis.
-
An assay using the robust homing potential of these cells reveals a ‘coherent feed-forward’ transcriptional subcircuit composed of Pax6, Six3, Six1/2, Eya, and Dach1 that is responsible for the directed homing mechanism of these multipotent progenitors.
-
Each of the five transcription factors involved in controlling homing is expressed in the aboral coelomic pouch. Transcription factors Dach1, Eya, Six1/2, and Pax6 (Fig. 7; middle) are co-localized in the aboral coelomic pouch (Luo et al., 2012).
-
Fig. 8 (bottom): Six3 positively regulates eya and pax6 in the coelomic pouch mesoderm. Pax6 positively regulates six3, eya, and itself.
-
References
Zhao et al. (2015)
Martik and McClay (2015)
Phb1 | Class: TF
-
Yamazaki et al. (2020): In euechinoids, Pmar1 promotes endomesoderm specification by repressing the hairy and enhancer of split C (hesC) gene. Here, we have identified the basal echinoid (cidaroid) pmar1 gene, which also promotes endomesoderm specification but not by repressing hesC. A further search for related genes demonstrated that other echinoderms have pmar1-related genes named phb. Functional analyses of starfish Phb proteins indicated that, similar to cidaroid Pmar1, they promote activation of endomesoderm regulatory gene orthologs via an unknown repressor that is not HesC.
-
Right (Fig. 2): All phb genes examined were expressed in the endomesoderm region at the vegetal pole. At the mid-gastrula stage (24 h), phbA expression was detected in the region encircling the blastopore (Fig. 2J), which seems to be endoderm lineage, but phbB expression was no longer detected (Fig. 2O)
-
Yamazaki et al. (2020).
SoxE | Class: GermLineDeterminant, TF
-
Juliano et al. (2006):
-
Top (Fig. 5): Sp-SoxE transcripts accumulate in the small micromere descendents at the tip of the archenteron in gastrula stage embryos and in the left coelomic pouch of the pluteus.
-
Sp-SoxE is a member of the HMG-box containing transcription factor family and the mouse homologs, Sox9 and Sox8, are expressed in the embryonic male gonad and are required for male sexual differentiation (Chaboissier et al., 2004, Kent et al., 1996). Therefore, the dimorphic expression pattern of Sp-SoxE in S. purpuratus plutei suggests a conserved sex-specific role. Furthermore, the localization of Sp-SoxE transcript in the left coelomic pouch supports the hypothesis that this is the site of germ cell accumulation. The left coelomic pouch is the future site of the adult rudiment.
-
-
Walton et al. (2009): Normal soxE expression is in the left coelomic pouch in L. variegatus as well.
References
Juliano et al. (2006)
SoxH | Class: TF
-
Abdelalim et al. (2014): Pluripotent stem cells (PSCs), including embryonic stem cells (ESCs), and induced PSCs (iPSCs) are able to self-renew and differentiate into a multitude of specialized cellular lineages. In these cells, the pluripotential identity is maintained by a group of transcription factors (TFs). Among these factors, SOX TFs play an essential role, not only in regulating pluripotency but also in mediating self-renewal and differentiation. Some SOX TFs are highly expressed in undifferentiated PSCs, while others are upregulated upon differentiation to promote specific lineage differentiation. Further roles of SOX factors in pluripotency are highlighted through their critical involvement in iPSCs generation. To perform these multiple functions and activities, SOX TFs are strongly associated with a complex regulatory network(s) that involves the binding of SOX factors to variant trans-acting partners to activate or suppress specific genes.
-
Little to no information about role and function in sea urchins/echinoderms available.
Tbr | Class: Immunity, TF
-
Wahl et al. (2009): The sea urchin regulatory gene tbrain (tbr) is zygotically expressed in the skeletogenic mesoderm (SM) of the cleavage and blastula stage embryo (Croce et al., 2001, Oliveri et al., 2002), and its expression is required for the postgastrular formation of the larval spicules (Fuchikami et al., 2002). Through transcriptional activation of a target gene, erg, tbr establishes an erg-hex-tgif-alx1 positive feedback circuit that maintains the regulatory state of the skeletogenic mesoderm domain from early in development and eventually, together with other regulators, serves as a transcriptional driver of an initial set of differentiation genes (Oliveri et al., 2008). The tbr gene thus has essential roles, first in specification of the SM and then in definitive larval skeletogenesis.
-
Top (Fig. 1C): GFP fluorescence image overlays of tbr::GFP BAC-injected embryos at 18, 24, and 48 hpf. Expression is limited to the skeletogenic cells at all stages:
-
Left (Fig. 9A): An updated tbrain regulatory subcircuit showing the inputs into the γ(2) module from Ets1/2 and Erg identified in this study. The diagram displays interactions that occur in the SM prior to ingression.
-
-
Rho and McClay (2011): Pmar1, Ets1 and Tbr are necessary for activation of foxN2/3 in micromeres.
References
Wahl et al. (2009)
Tbx4 | Class: TF
-
NCBI: Tbx4 is a member of a phylogenetically conserved family of genes that share a common DNA-binding domain, the T-box. T-box genes encode TFs involved in the regulation of developmental processes. Expression studies in mouse and chicken show that Tbx4 is expressed in developing hindlimb, but not in forelimb buds, suggesting a role for this gene in regulating limb development and specification of limb identity.
-
Other T-box TFs in sea urchins include ske-T (expressed in skeletogenic mesenchyme lineage; Croce et al. 2001), LvTbx2/3 (involved in formation of O/A axis; Gross et al. 2003), and Coquillette (expressed asymmetrically along the O/A axis and involved in skeletogenesis; Croce et al. 2003).
References
Oliveri and Davidson (2004); Fig. 3: GRN for sea urchin endomesoderm specification: the view from the genome. The architecture of the network is based on perturbation and expression data, and cis-regulatory analyses.
Figure 2.
Figure 2. Gene expression profiles of transcription factors of interest in S. purpuratus embryos over time. (A) Line plots illustrating the transcripts per embryo as a percentage of maximum expression levels for each gene over time. (B) Line plot illustrating transcripts per embryo for each of the nine transcription factors of interest. FoxA (lime green) consistently has the highest relative expression throughout development. Data were obtained from the Echinobase online resource (http://legacy.echinobase.org/shiny/quantdev/). Note that data represent transcripts per entire embryo and not from a specific embryonic territory.
A.

B.

Figure 3.
Figure 3. Transcription factors of interest in order of activation. Nine transcription factors of interest identified using the methods described in Sections 2.2 and 2.3 ordered sequentially according to gene expression profile seen in Fig. 2. Transcription factors are listed at timepoints when their expression peaks. At the gastrula stage during archenteron formation (30 hours post-fertilization) FoxA expression is at its peak.

Putting it all together


































